ONT Fiber-seq Analysis
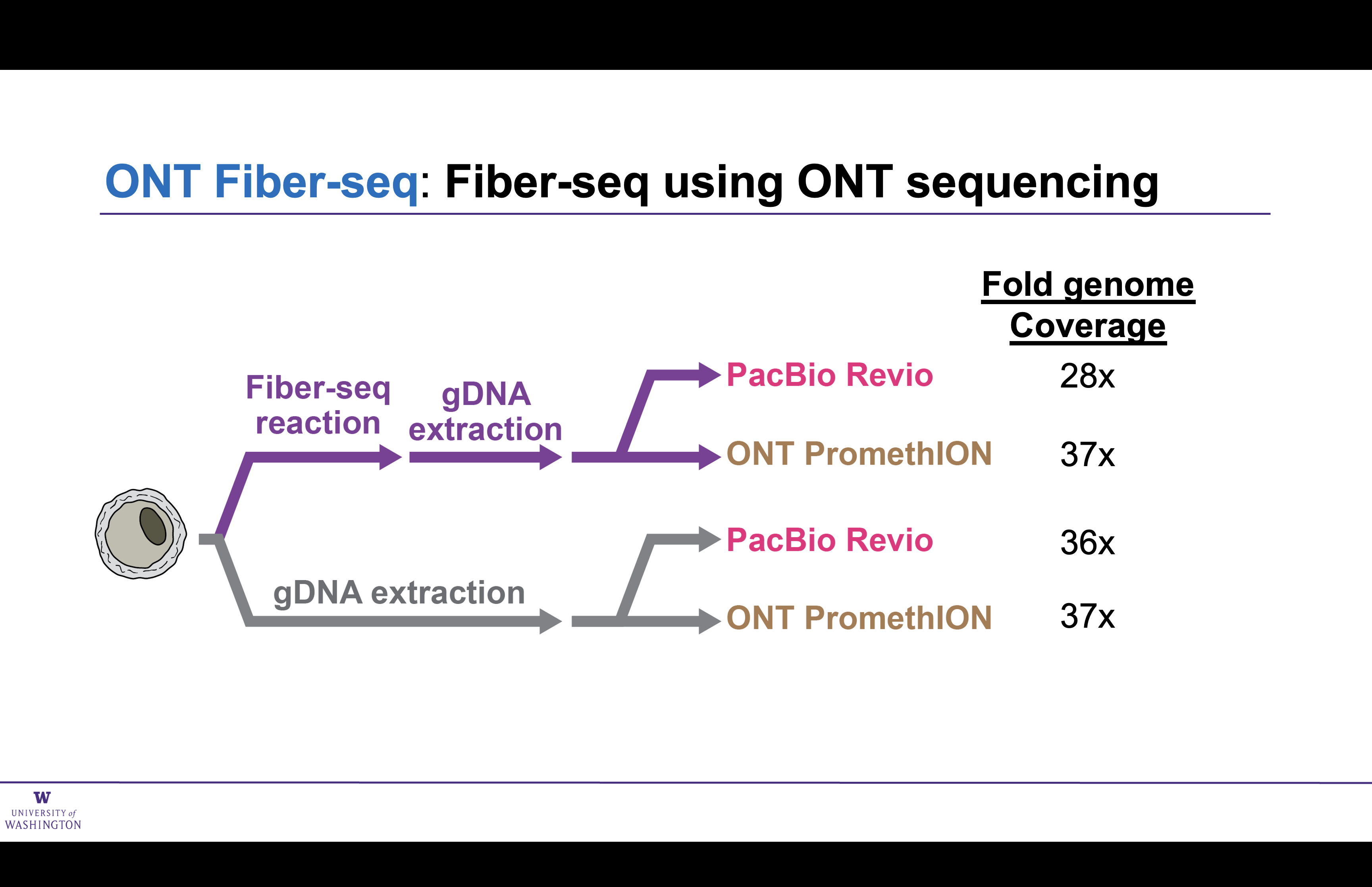
Matched ONT and PacBio preparations
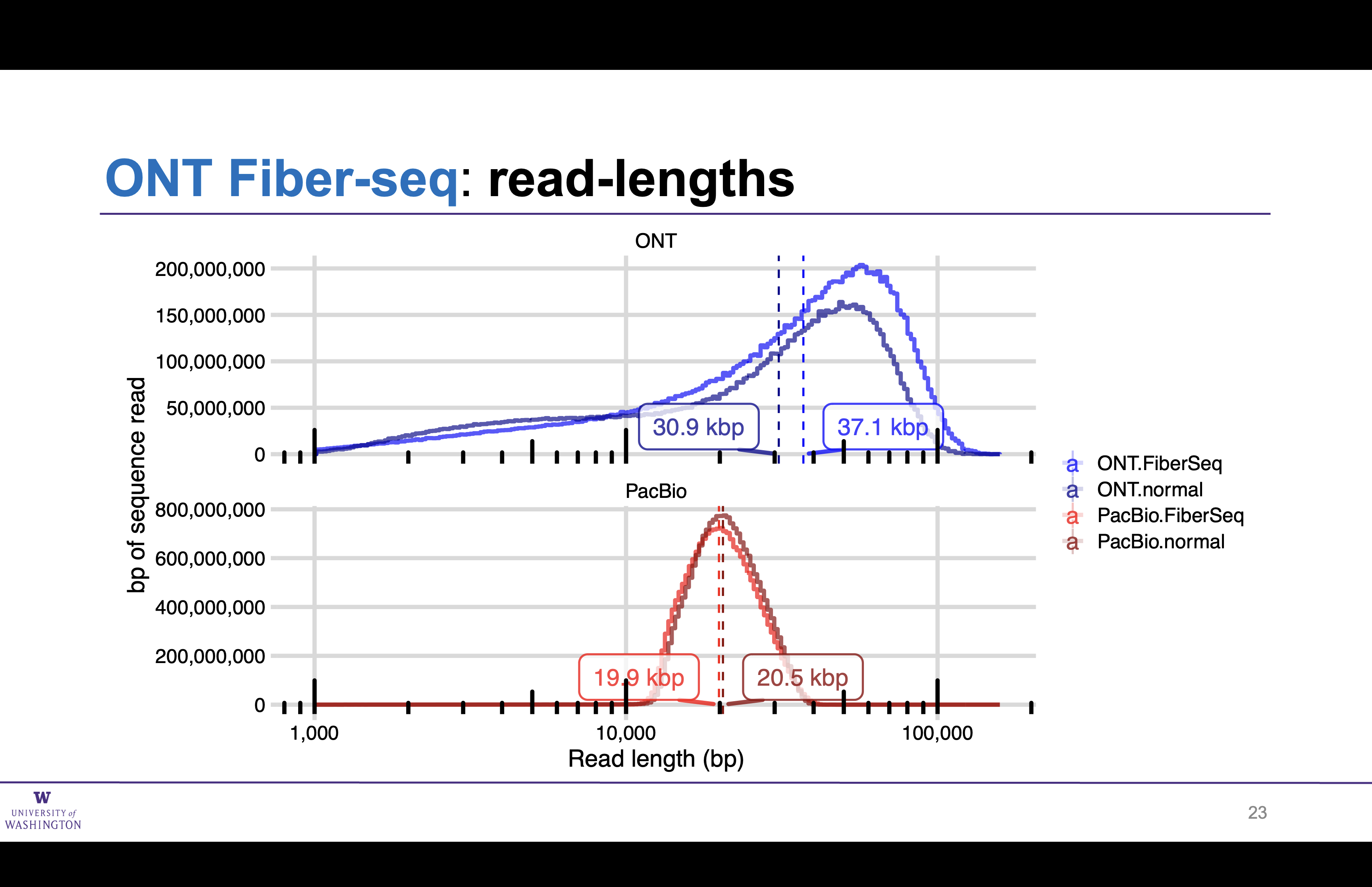
Read lengths unchanged by Fiber-seq
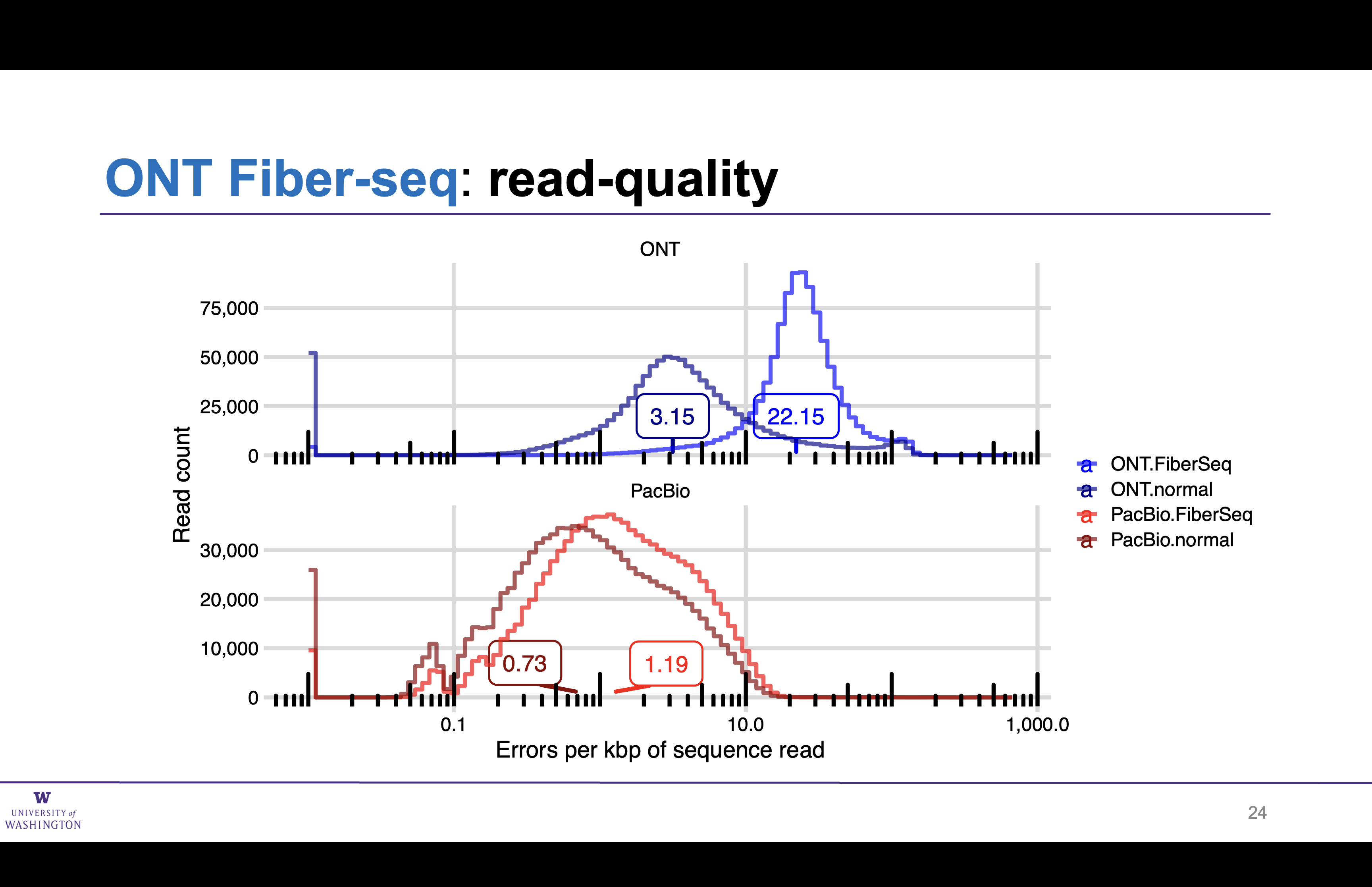
Impact of Fiber-seq on ONT read quality
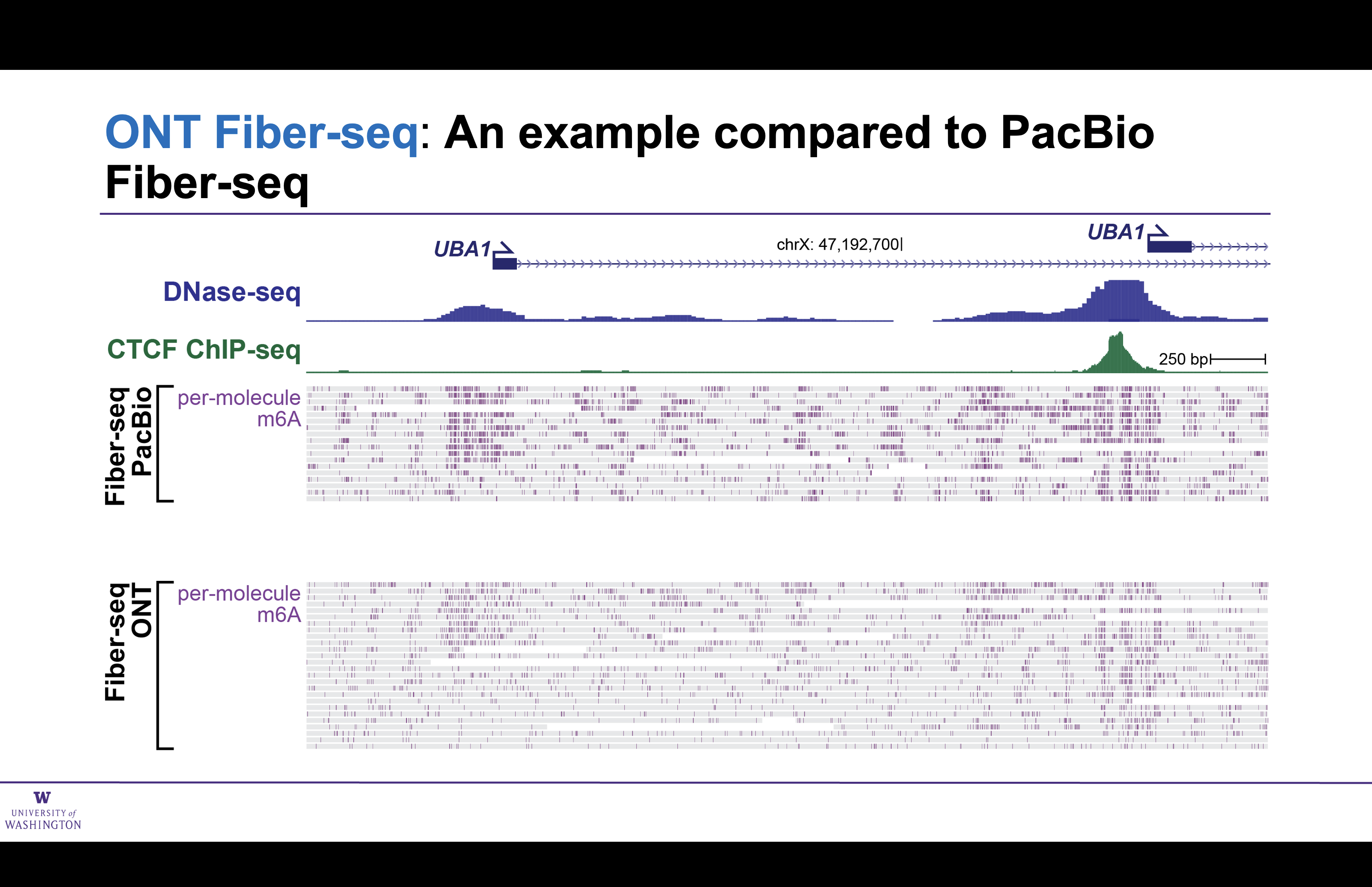
A comparison of ONT and PacBio Fiber-seq
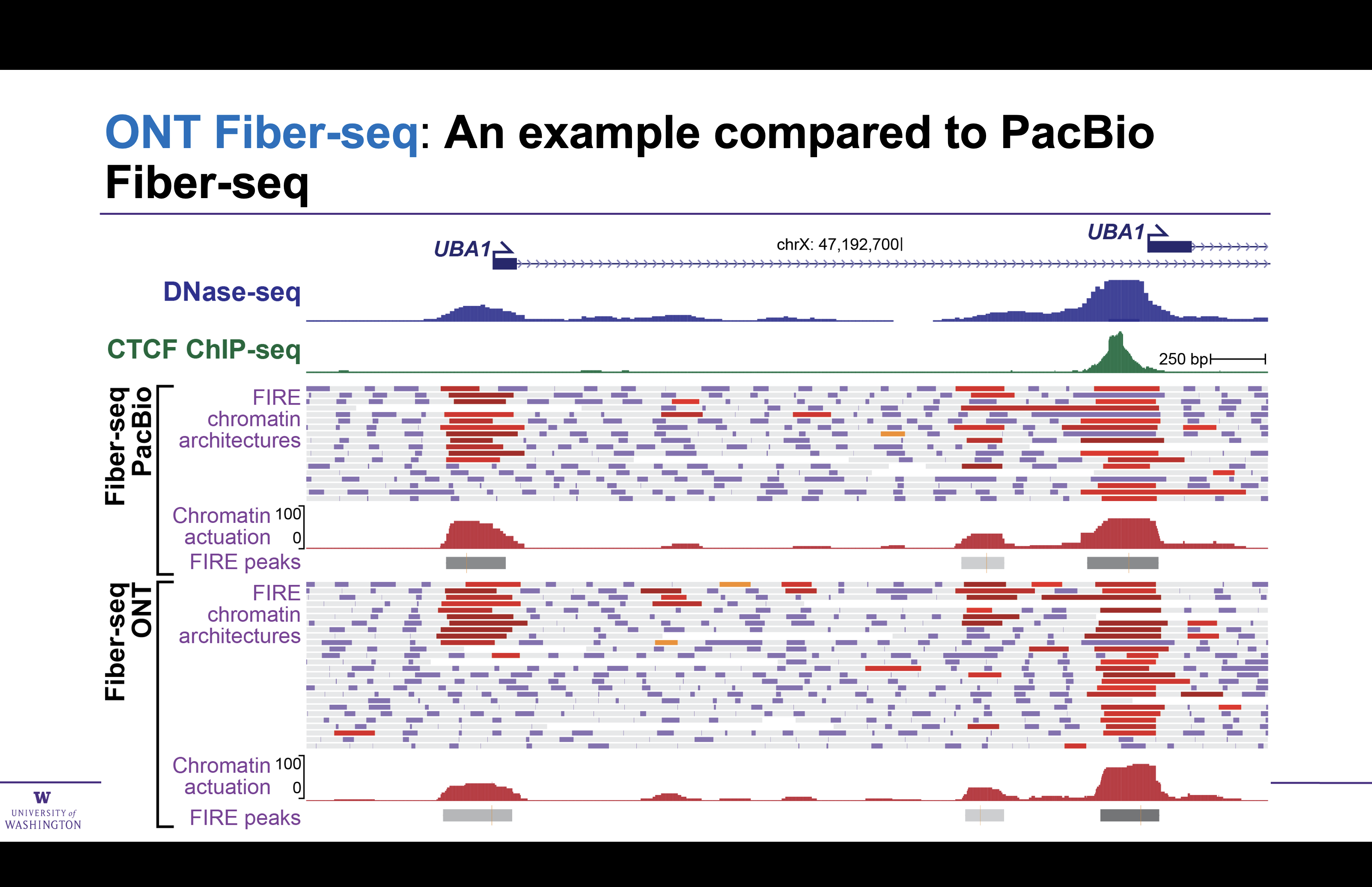
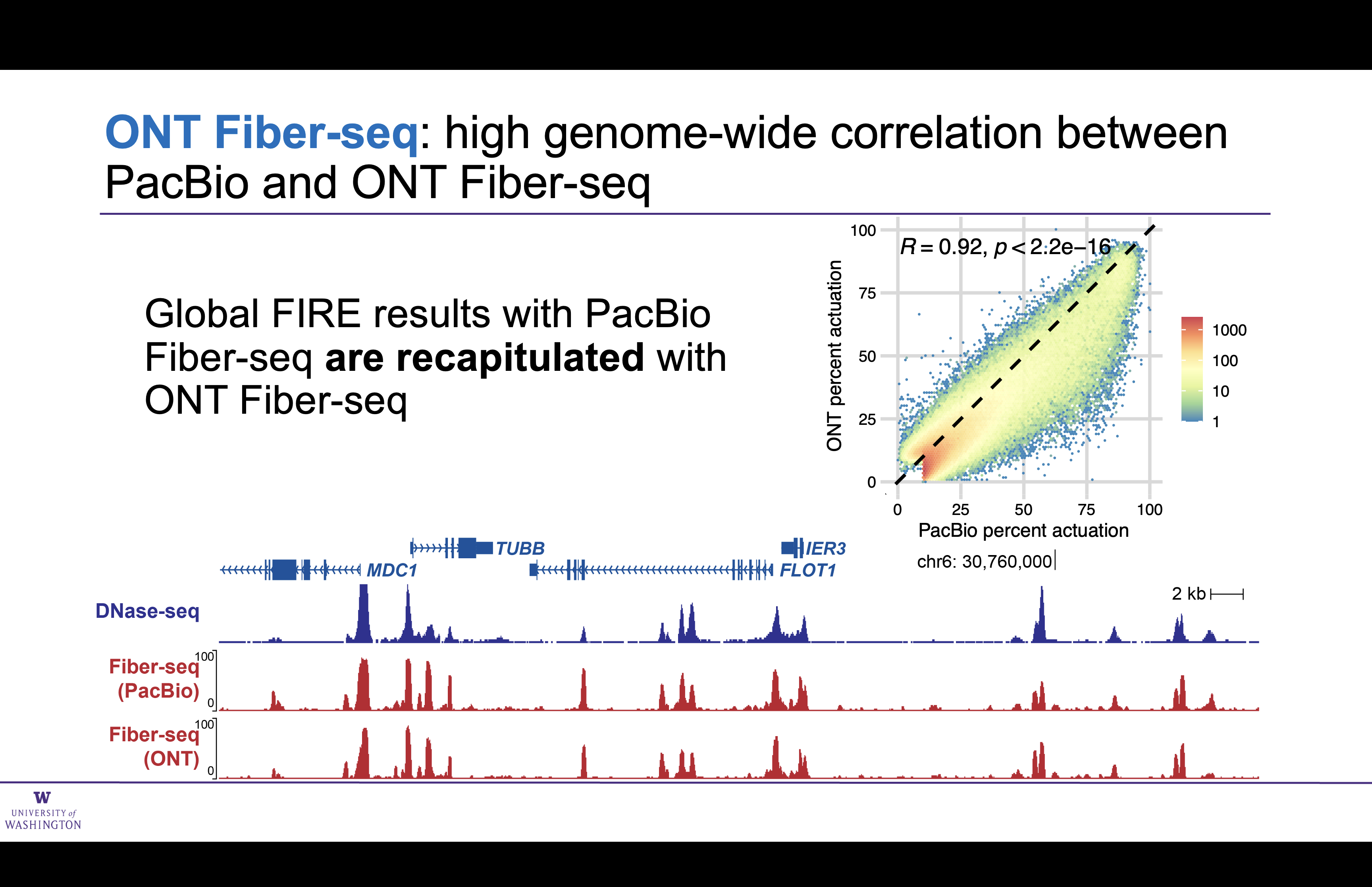
Genome-wide comparison of ONT and PacBio Fiber-seq
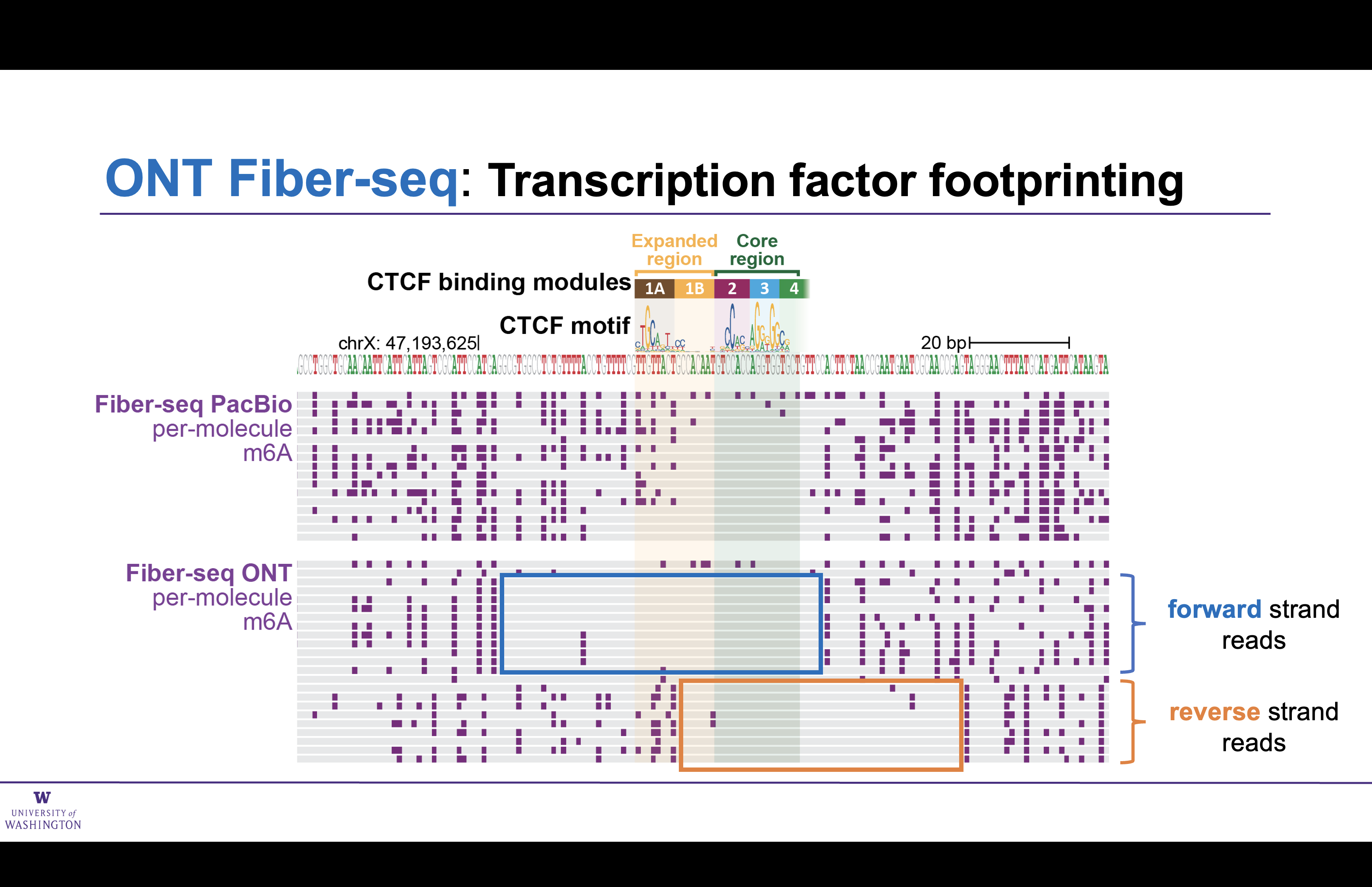
Transcription factor foot printing has reduced resolution with ONT
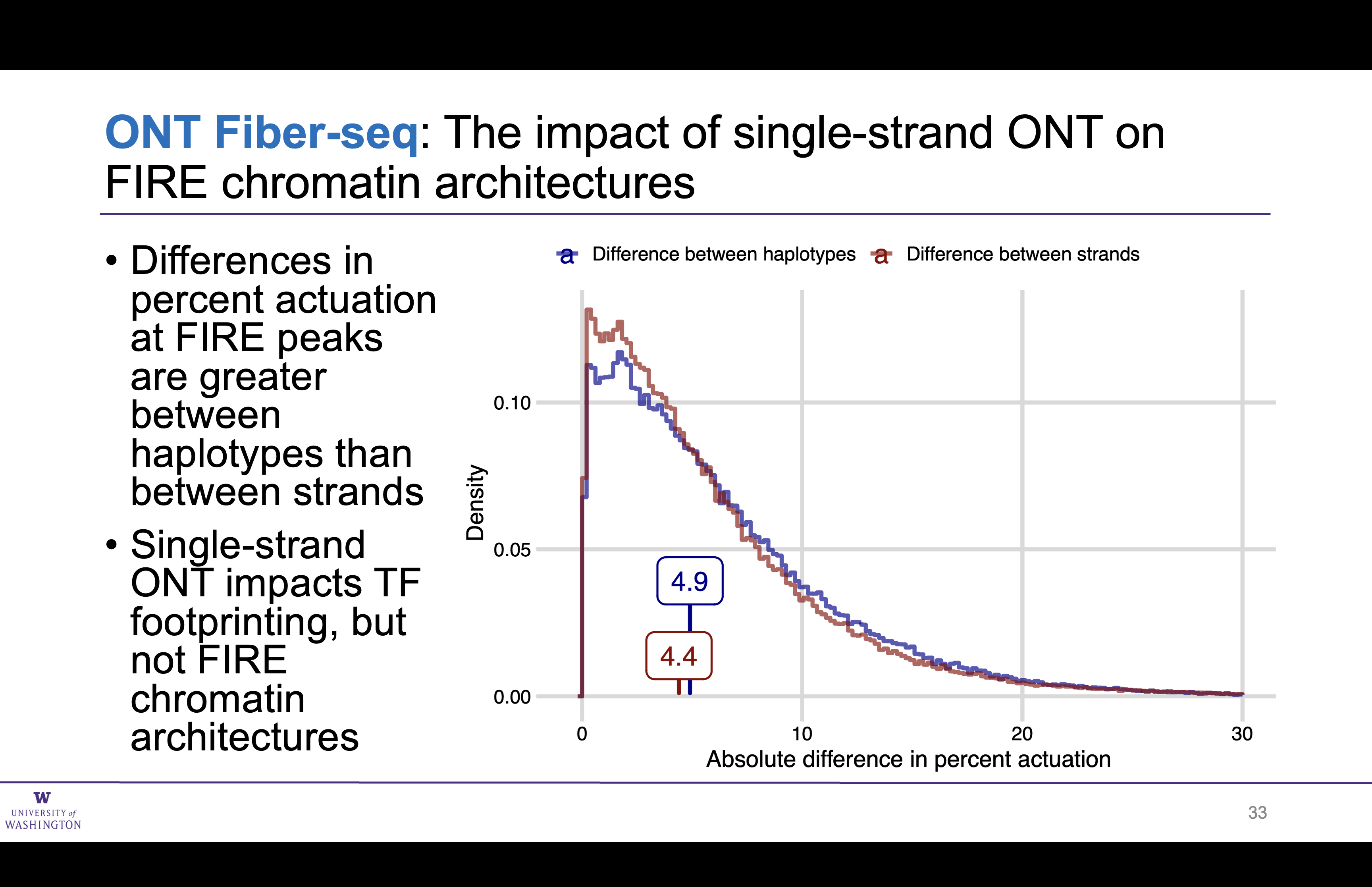
Stand does not significantly impact ONT Fiber-seq FIRE results
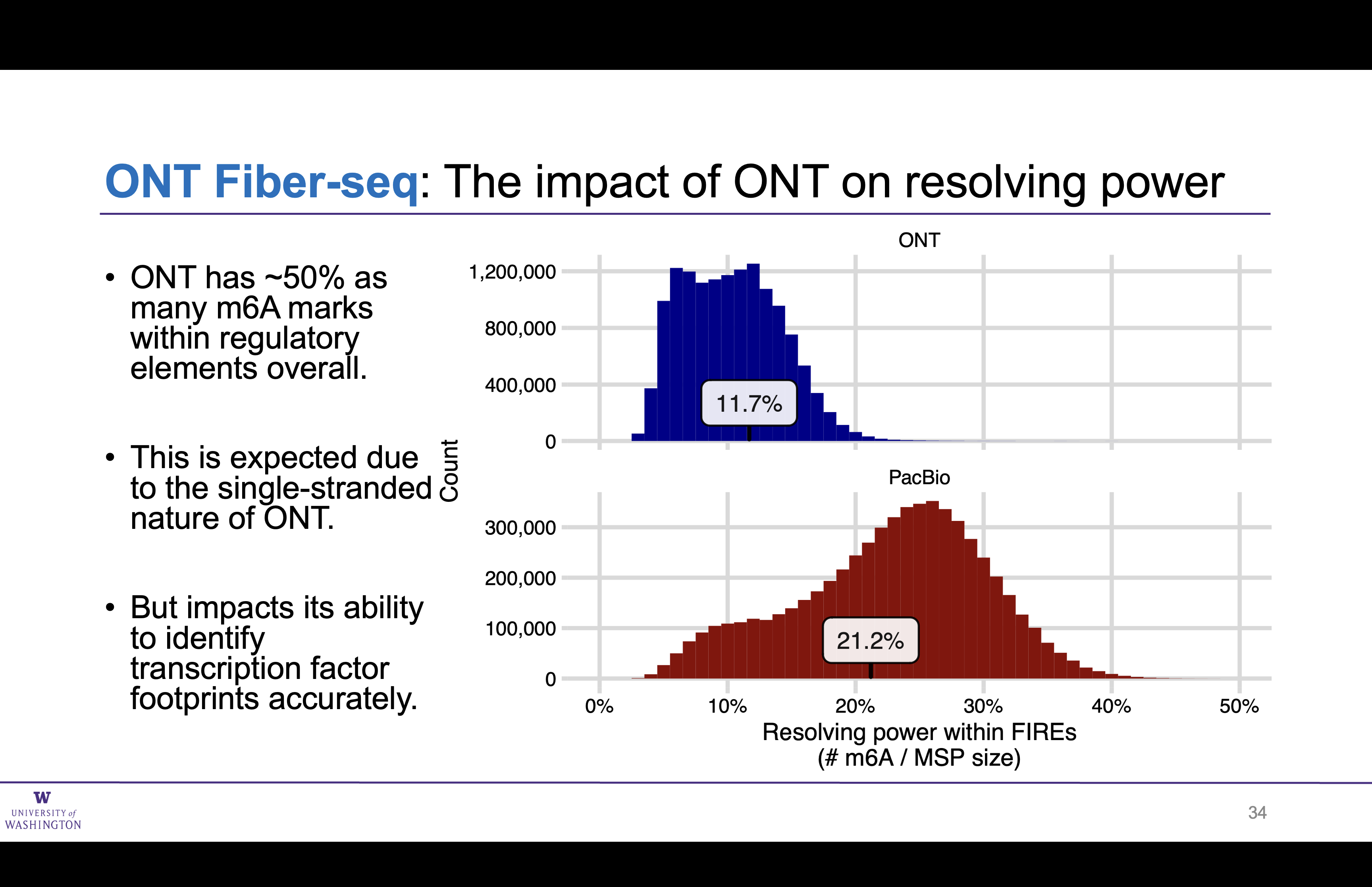
ONT FIREs have ~half as many m6A sites as PacBio (as expected)
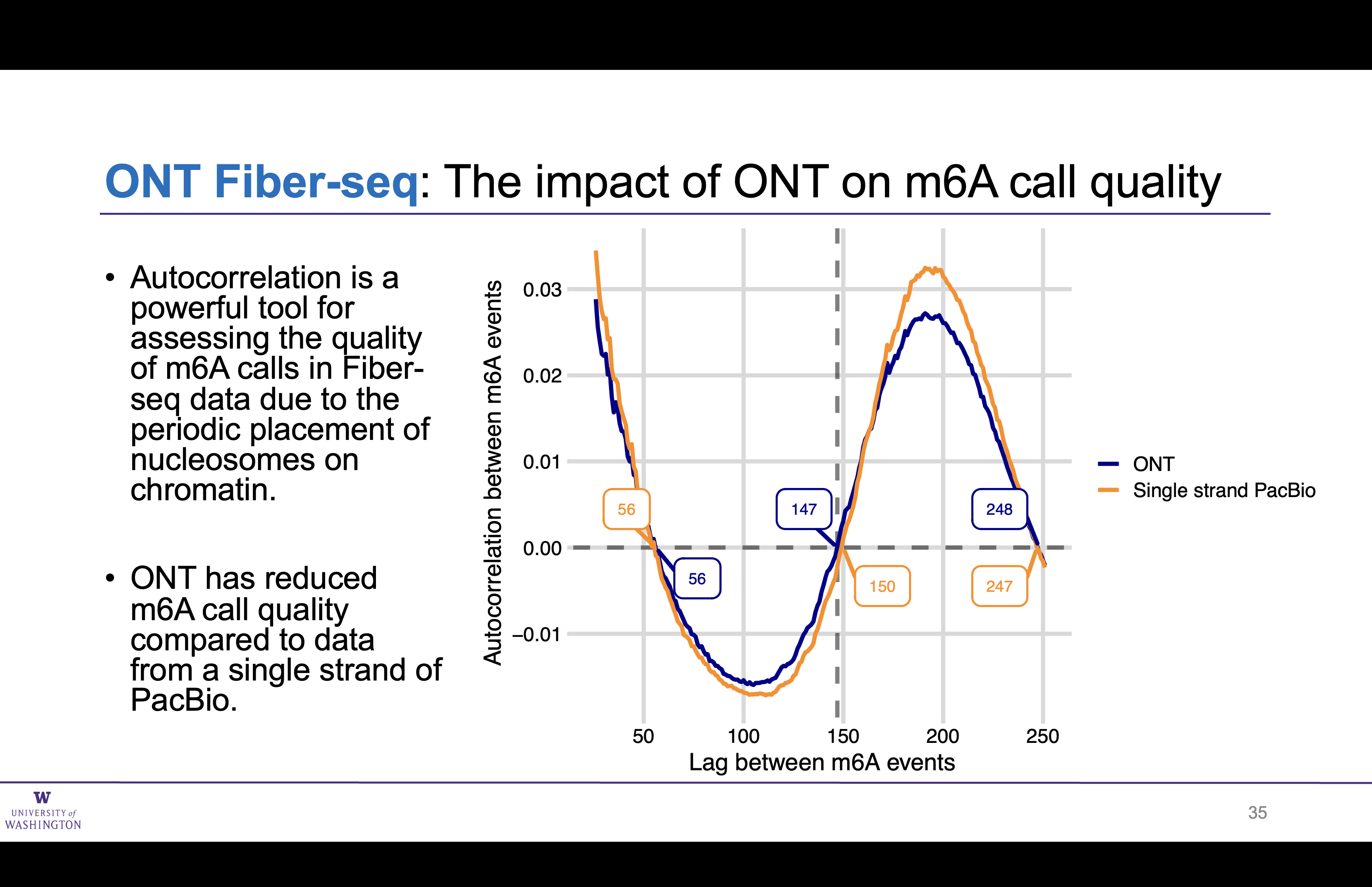
Auto-correlation of m6A sites in ONT nearly matches a single strand of PacBio
Overall conclusions